Pre-Alcohol
A probiotic that digests a byproduct of alcohol – built with genetic engineering by a team of microbiologists.
The Problem
Acetaldehyde is an unwanted molecule that can build up in the body when drinking alcohol
Here’s the problem. The body has many biological reactions to drinking alcohol, not all of which are pleasant. The metabolism of alcohol produces a variety of small molecules and cellular responses which, when piled-up, can be hard on your body and throw you off your healthy routine. One of those small molecules is acetaldehyde, an unwanted, toxic byproduct of alcohol metabolism. Its chemical structure is highly reactive, which means it can interact with a whole host of molecular and cellular components throughout our bodies. But how does alcohol become acetaldehyde?

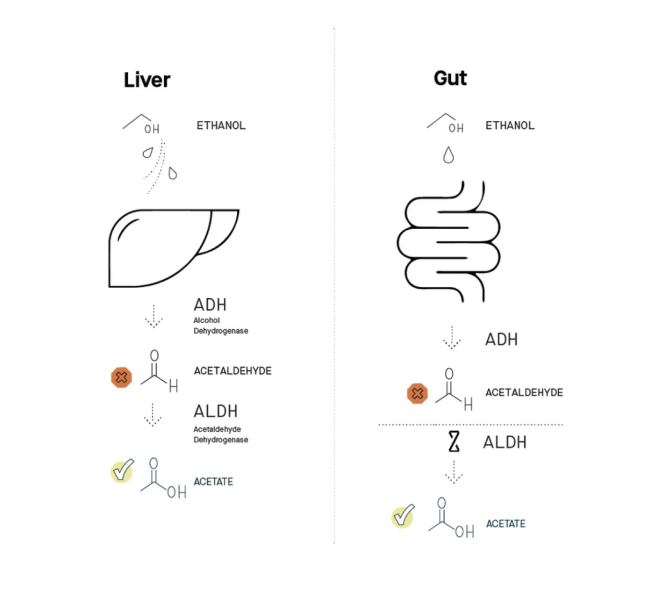
The liver can fully metabolize acetaldehyde, but the gut isn’t properly equipped to do the same
When you drink, most of the alcohol you consume gets digested in the liver using a two-step process (see diagram). Each step requires a different functional molecule produced by your liver – functional molecules called enzymes. First, alcohol is converted into acetaldehyde with an enzyme called alcohol dehydrogenase (ADH). Second, acetaldehyde is converted into another chemical called acetate with an enzyme called acetaldehyde dehydrogenase (ALDH). That final product, acetate, is benign (it’s essentially vinegar). Via this two-step process, alcohol in your liver is rapidly metabolized into harmless acetate.
However, some of the alcohol you drink never reaches your liver. Instead, it is metabolized in your gut, in large part by the microbes that reside there. Some microbes in the gut microbiome are equipped with the first enzyme, ADH, and can convert alcohol into acetaldehyde. But in contrast to your liver, they don’t make enough of the second enzyme, ALDH, to convert acetaldehyde into acetate. This missing trait is the major source of acetaldehyde buildup in your body (citation).
Thus arises a clear hypothesis: If we can equip the gut microbiome with the ability to break down acetaldehyde into acetate, then we can augment the body’s ability to minimize the buildup of this toxic byproduct. That is a result that would be very beneficial for those planning to have a drink or two. The liver can fully metabolize acetaldehyde, but the gut isn’t properly equipped to do the same.
The Solution
Probiotics can be engineered to break down acetaldehyde in the gut
If you’re trying to enable the gut microbiome to break down acetaldehyde, the most straightforward approach would be to make a bunch of the same enzyme your liver uses (ALDH) and deliver it to the gut, mimicking exactly what your body already does so effectively. Unfortunately, in this case (and most cases really) we can’t just eat an enzyme to equip our bodies with new traits. Enzymes are proteins, and most proteins you consume will break down very quickly as food in your stomach. Once broken down they lose their function.
But our scientists knew three very interesting things:
- There are probiotic bacteria that can survive the harsh environment of the stomach unharmed.
- Many bacteria are capable of making ALDH – the very enzyme our livers use to break down acetaldehyde.
- We can use genetic engineering to create a probiotic bacteria with the ability to make ALDH!
So rather than trying to solve the problem with an enzyme that won’t survive in the gut, we can instead use genetic engineering to build a probiotic bacteria that not only makes the enzyme but can also deliver it safely to where it's most needed to break down acetaldehyde!
Though the actual building and testing behind our engineered ZB183™ strain took years, here is what we did on a basic level: We started with a natural probiotic bacteria called Bacillus subtilis. Humans have been intentionally consuming B. subtilis as a probiotic in supplements and fermented foods (e.g. natto, kombucha, etc.) for centuries. B. subtilis also has the natural ability to pass through your stomach acid unharmed and make enzymes in your gut (citation), We then used a process naturally honed by bacteria to insert the instructions for making ALDH into the bacteria’s genome, carefully selecting a location that optimizes the expression of this enzyme. The result was ZB183™ – a probiotic strain of B. subtilis that is identical in every way to the one humans already consume, but with one additional trait: the ability to break down acetaldehyde.
Data & Validation
Our engineered probiotic has been tested in the lab for safety and functionality
Once ZB183™ was built, we needed to ensure that it was safe, and that it worked as intended.
Safety
We subjected ZB183™ to years of laboratory testing and review by America’s top food toxicologists, the results of which confirmed it to be completely safe and FDA-compliant. The data from these tests has been published in the peer-reviewed Journal of Toxicology.
Now that we have confirmed our genetically engineered probiotic is safe and works as intended, we make sure this is true of every batch. Pre-Alcohol undergoes rigorous quality testing at every stage, from probiotic biomass to the final product, ensuring safety, purity, and efficacy. We transparently publish our standards for each test on our Quality page which can be found here.
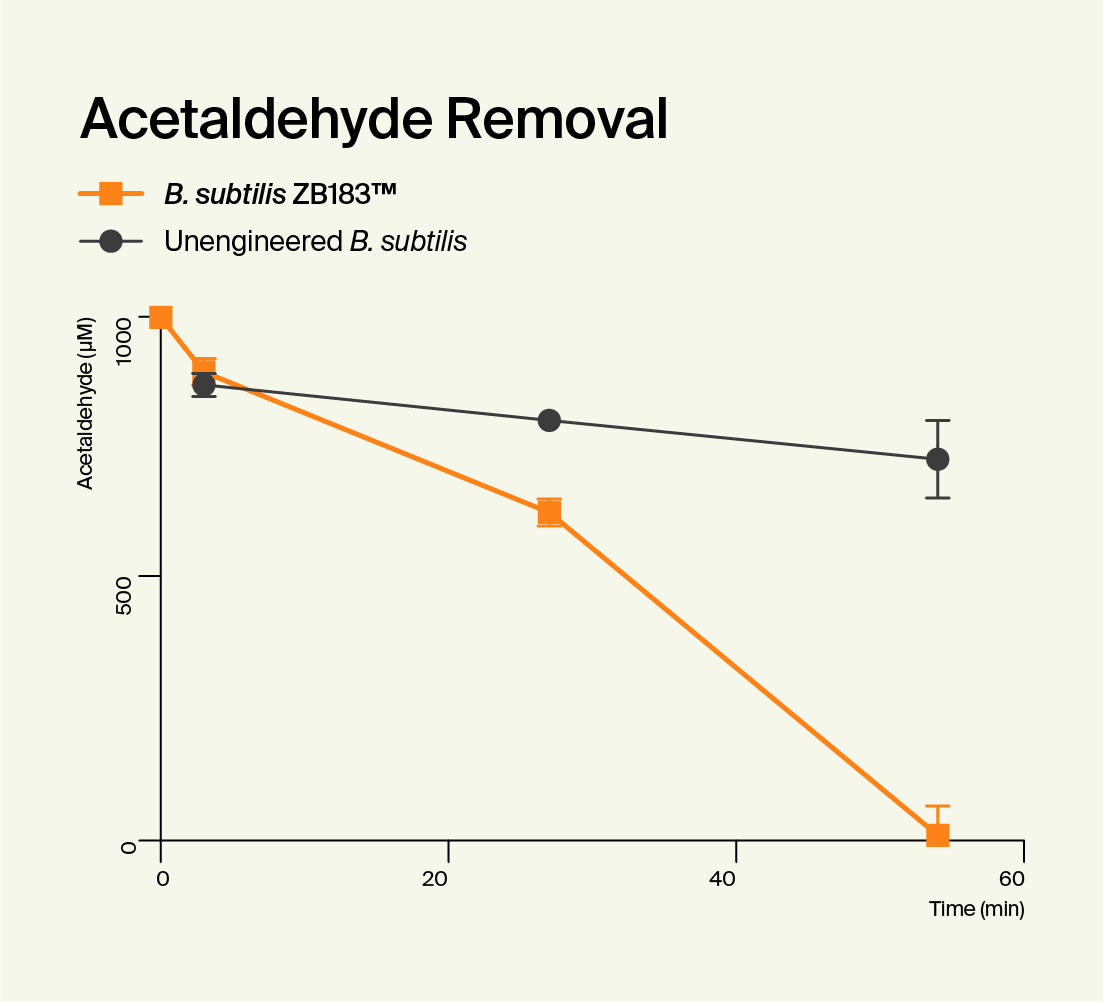
Functionality
To confirm that ZB183™ worked as intended, we tested its ability to make ALDH and break down acetaldehyde. Over a series of laboratory tests where we compared it to the unedited B. subtilis strain, we confirmed that our Pre-Alcohol Probiotic was highly effective at producing ALDH. We also confirmed that while the control strain broke down almost no acetaldehyde, ZB183™ was able to break down more acetaldehyde than you could ever encounter when drinking. We published a paper containing the data from these tests, and it can be found in the peer-reviewed PLOS One journal..
As scientists we appreciate that every body is different and so there are no silver bullets in biology. Even though we’ve proven that the product can reliably provide the function of helping to metabolize acetaldehyde, we don’t know for sure that it will be perceived as valuable for everyone. However, we do know from customer use data that it does appear to provide a benefit to the vast majority of users (i.e. >95%). And with millions of bottles sold, that’s a lot of happy customers!
ZB183™ was our first proof of concept on a bigger journey to unlock the immense potential of safely and consciously-built genetically engineered microbes.
Founded by Scientists
Who are we? The very same people who did the research and built this technology: the ZBiotics team. We pride ourselves on the fact that we are a team founded and led with science at our core. Our technology is developed in-house by our own scientists in our own microbiology lab. This integrated approach allows us to create the best products available — products that marry cutting edge technology with innovative product design.
Our founder, CEO, and inventor of the primary technology underlying Pre-Alcohol is Dr. Zack Abbott. Zack’s background is as a microbiologist; he received his PhD in Microbiology & Immunology from the University of Michigan. After that, he worked designing clinical trials for biotechnology and pharmaceutical companies prior to starting ZBiotics to bring the idea of genetically engineered probiotics to the world.
To learn more, read about our founding story or meet the team.
You probably have some questions
Check out our FAQs to find answers to common questions and feel free to reach out if you have more.
When you're drinking. The earlier the better.
The probiotic in Pre-Alcohol needs time to “wake up” in your gut, so we say the earlier the better if you can remember it. We drink Pre-Alcohol just before going out – right before our first drink. But it should be active if you drink it any time during the day or evening. So whether it’s a few hours before drinking, right with your first drink, or even if you’ve already enjoyed a round or two, go for it.
The probiotic in Pre-Alcohol is likely to stay in your body for 18-24 hours after you drink it. So one bottle will cover you for a whole day, even if you're starting with brunch mimosas followed by evening plans later on!
Yes. Pre-Alcohol is fully FDA-compliant for safety and adheres to all regulatory requirements for sale in the U.S.
Note that FDA compliance is not FDA approval. FDA approval is a process reserved for drug products. Pre-Alcohol is a food/beverage product and has not been approved as efficacious for any purpose by the FDA.
FDA-compliant for safety means that we’ve satisfied all regulatory requirements to ensure Pre-Alcohol is safe and can be legally sold in the United States. That means that all our manufacturing is conducted in an FDA-registered food-grade facility according to Good Manufacturing Practices compliant with 21 CFR 117. It also means that our novel ingredient – Bacillus subtilis ZB183™ – has been extensively tested and is generally recognized, among qualified experts, as having been adequately shown to be safe under the conditions of its intended use.
No.
Pre-Alcohol will not affect your level of intoxication. Pre-Alcohol does not decrease your blood alcohol level, nor does it make alcohol any safer.
All the same rules about alcohol safety apply. Don’t get behind the wheel, don't operate heavy machinery, and don't drink too much. Alcohol is still dangerous on its own (citation).
That's not how it's designed.
We intentionally designed Pre-Alcohol so that it would not change your microbiome. We purposefully built our probiotic using a base bacteria that is well-adapted to passing straight through you and not colonizing your gut.
This creates a few likely benefits: (1) your microbiome remains unchanged; (2) the interaction between our probiotic and your gut should remain consistent and predictable; and (3) because we’re not trying to change your microbiome, we don’t need to use an excessive amount of probiotic per serving.